Pre-processing of new datasets
09 July, 2020
Last updated: 2020-07-09
Checks: 7 0
Knit directory: Epilepsy19/
This reproducible R Markdown analysis was created with workflowr (version 1.6.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200706) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0c778bb. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/fig_go.nb.html
Ignored: analysis/fig_neun.nb.html
Ignored: analysis/fig_overview.nb.html
Ignored: analysis/fig_smart_seq.nb.html
Ignored: analysis/fig_summary.nb.html
Ignored: analysis/fig_type_distance.nb.html
Ignored: analysis/gene_testing.nb.html
Ignored: analysis/prep_alignment.nb.html
Ignored: analysis/prep_filtration.nb.html
Ignored: cache/con_allen.rds
Ignored: cache/con_filt_cells.rds
Ignored: cache/con_filt_samples.rds
Ignored: cache/con_ss.rds
Ignored: cache/count_matrices.rds
Ignored: cache/p2s/
Ignored: output/
Untracked files:
Untracked: analysis/figure/
Untracked: code/
Untracked: data_mapping.yml
Unstaged changes:
Modified: README.md
Modified: analysis/index.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/prep_filtration.Rmd) and HTML (docs/prep_filtration.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 0c778bb | viktor_petukhov | 2020-07-09 | metadata and python path |
| Rmd | 83671cf | viktor_petukhov | 2020-07-09 | Pre-processing notebooks |
# library(Epilepsy19)
library(ggplot2)
library(magrittr)
library(Matrix)
library(pbapply)
library(conos)
devtools::load_all()
theme_set(theme_bw())Load data:
cms <- list.files(DataPath(""), pattern="*.h5") %>% setNames(., gsub(".h5", "", .)) %>%
pblapply(function(p) DataPath(p) %>% Seurat::Read10X_h5() %>% .[, colSums(.) > 10])Filter small cells:
thresholds <- c(Biopsy=3.05, GTS213=2.5, GTS217=1.0, GTS217_2=3.2, GTS219=2.5, GTS233=3.0,
HB26=3.4, HB51=3.2, HB52=2.85, HB53=3.1, HB56=2.75, HB65=2.85, NeuN=3.0,
CTR215=2.7, CTR240=3.0)
lapply(names(cms), function(n)
dropestr::PlotCellsNumberHist(colSums(cms[[n]]), estimate.cells.number=T, show.legend=F) +
geom_vline(aes(xintercept=thresholds[[n]]))) %>%
cowplot::plot_grid(plotlist=., ncol=3, labels=names(cms))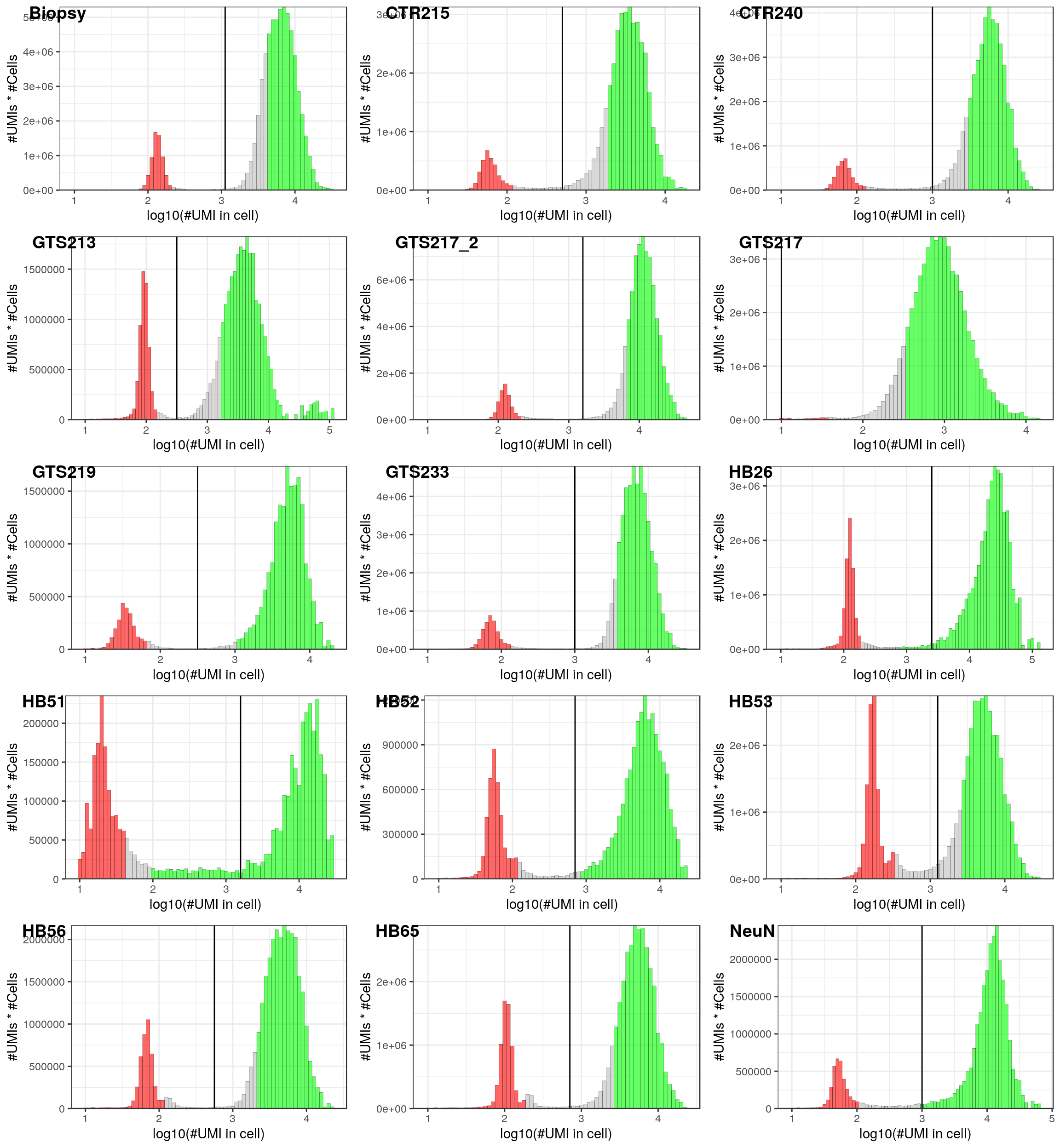
cms <- names(cms) %>% setNames(., .) %>%
pblapply(function(n) cms[[n]] %>% .[, colSums(.) >= 10**thresholds[[n]]])Append matrices from the previous runs:
cm_paths_old <- list.files(DataPath(""), pattern=".*_p.*") %>%
setNames(., .) %>% sapply(DataPath)
cms_old <- pbapply::pblapply(cm_paths_old, pagoda2::read.10x.matrices, verbose=F, cl=10)
cms %<>% c(cms_old)
sapply(cms, ncol) Biopsy CTR215 CTR240 GTS213 GTS217_2 GTS217 GTS219 GTS233
10661 12726 9491 8859 7475 106648 4508 8030
HB26 HB51 HB52 HB53 HB56 HB65 NeuN c_p1
2314 283 3184 7517 6806 7046 2822 4693
c_p2 c_p3 c_p4 ep_p1 ep_p2 ep_p3
1382 4857 3323 11304 6247 6390 sapply(cms, ncol) %>% .[!names(.) %in% c("GTS217", "NeuN")] %>% sum()[1] 127096Mitochondrial fraction:
mit_frac_per_dataset <- pblapply(cms, function(cm)
colSums(cm[grep("MT-", rownames(cm)), ]) / colSums(cm))lapply(mit_frac_per_dataset, function(fr)
qplot(fr[fr < 0.5], xlab="Mit. fraction", ylab="#Cells", xlim=c(-0.01, 0.5), bins=30)) %>%
cowplot::plot_grid(plotlist=., ncol=4, labels=names(cms))Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).
Warning: Removed 2 rows containing missing values (geom_bar).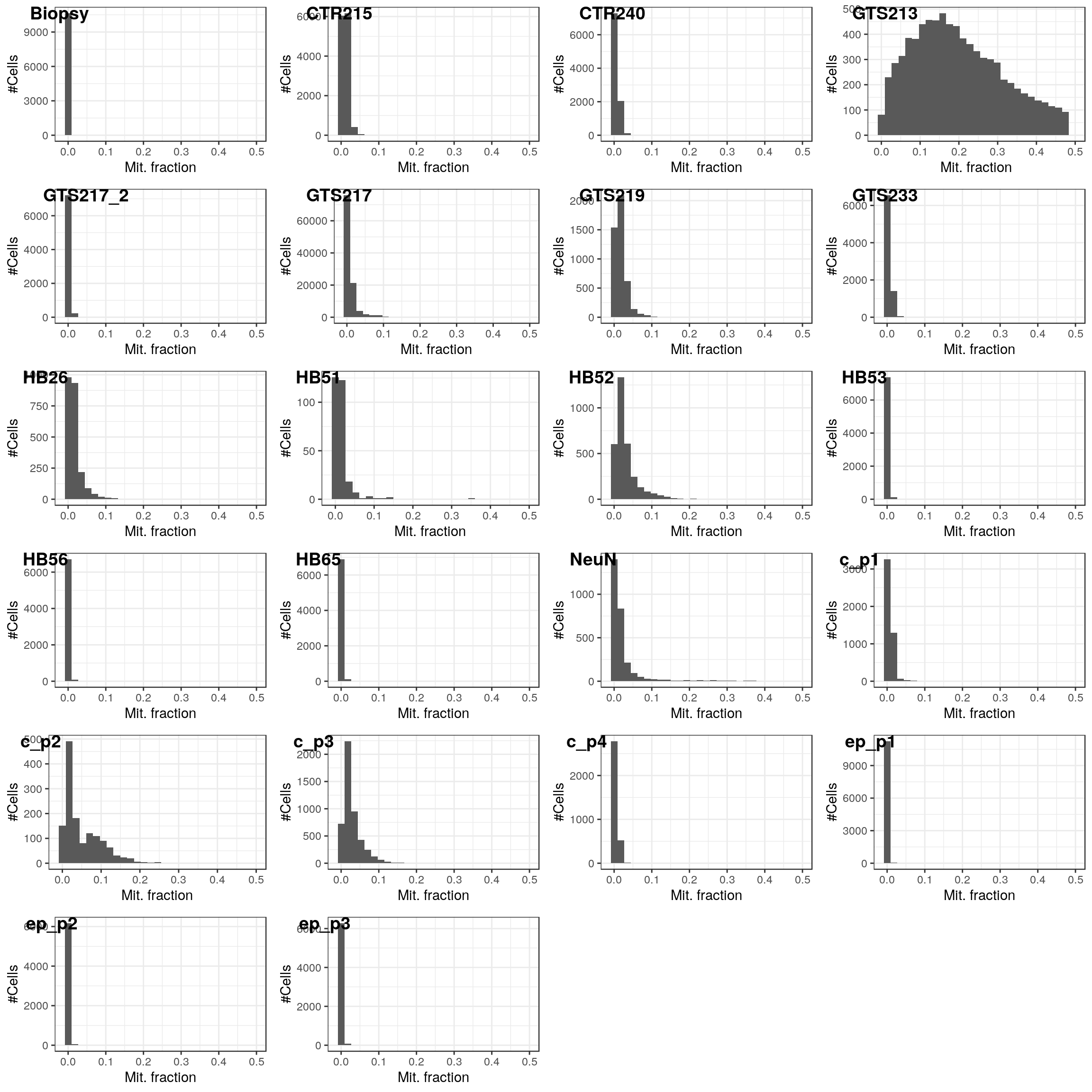
cms <- pbmapply(function(cm, frac) cm[, frac < 0.08], cms, mit_frac_per_dataset) %>%
setNames(names(cms))
cms$GTS213 <- NULL
cms$GTS217 <- NULL
sapply(cms, ncol) Biopsy CTR215 CTR240 GTS217_2 GTS219 GTS233 HB26 HB51
10661 12720 9490 7474 4458 8023 2268 277
HB52 HB53 HB56 HB65 NeuN c_p1 c_p2 c_p3
2926 7517 6801 7042 2600 4675 1027 4601
c_p4 ep_p1 ep_p2 ep_p3
3323 11302 6246 6390 Doublet detection:
doublet_info <- pblapply(cms, GetScrubletScores, "~/mh/local/anaconda3/bin/python3.7",
min.molecules.per.gene=50, cl=25)is_doublet <- lapply(doublet_info, `[[`, "score") %>% sapply(`>`, 0.25)
sapply(is_doublet, mean) %>% round(3) Biopsy CTR215 CTR240 GTS217_2 GTS219 GTS233 HB26 HB51
0.076 0.078 0.056 0.047 0.031 0.043 0.037 0.040
HB52 HB53 HB56 HB65 NeuN c_p1 c_p2 c_p3
0.035 0.044 0.046 0.041 0.022 0.034 0.049 0.036
c_p4 ep_p1 ep_p2 ep_p3
0.070 0.058 0.039 0.051 Filter matrices
cms_filt <- cms
for (n in names(cms_filt)) {
cms_filt[[n]] %<>% .[!grepl("^MT-", rownames(.)), !is_doublet[[n]]]
}
sapply(cms_filt, dim) Biopsy CTR215 CTR240 GTS217_2 GTS219 GTS233 HB26 HB51 HB52 HB53 HB56
[1,] 33681 33681 33681 33681 33681 33681 33681 33681 33681 33681 33681
[2,] 9852 11727 8954 7124 4318 7674 2183 266 2823 7190 6486
HB65 NeuN c_p1 c_p2 c_p3 c_p4 ep_p1 ep_p2 ep_p3
[1,] 33681 33681 33681 33681 33681 33681 33681 33681 33681
[2,] 6752 2542 4518 977 4437 3090 10647 6002 6063sapply(cms_filt, ncol) %>% .[names(.) != "NeuN"] %>% sum()[1] 111083Save data
readr::write_rds(cms_filt, CachePath("count_matrices.rds"))
data.frame(value=unlist(sessioninfo::platform_info()))| value | |
|---|---|
| version | R version 3.5.1 (2018-07-02) |
| os | Ubuntu 18.04.2 LTS |
| system | x86_64, linux-gnu |
| ui | X11 |
| language | (EN) |
| collate | en_US.UTF-8 |
| ctype | en_US.UTF-8 |
| tz | America/New_York |
| date | 2020-07-09 |
as.data.frame(sessioninfo::package_info())[c('package', 'loadedversion', 'date', 'source')]| package | loadedversion | date | source | |
|---|---|---|---|---|
| AnnotationDbi | AnnotationDbi | 1.44.0 | 2018-10-30 | Bioconductor |
| ape | ape | 5.3 | 2019-03-17 | CRAN (R 3.5.1) |
| assertthat | assertthat | 0.2.1 | 2019-03-21 | CRAN (R 3.5.1) |
| backports | backports | 1.1.5 | 2019-10-02 | CRAN (R 3.5.1) |
| base64enc | base64enc | 0.1-3 | 2015-07-28 | CRAN (R 3.5.1) |
| beeswarm | beeswarm | 0.2.3 | 2016-04-25 | CRAN (R 3.5.1) |
| bibtex | bibtex | 0.4.2.2 | 2020-01-02 | CRAN (R 3.5.1) |
| Biobase | Biobase | 2.42.0 | 2018-10-30 | Bioconductor |
| BiocGenerics | BiocGenerics | 0.28.0 | 2018-10-30 | Bioconductor |
| bit | bit | 1.1-15.2 | 2020-02-10 | CRAN (R 3.5.1) |
| bit64 | bit64 | 0.9-7 | 2017-05-08 | CRAN (R 3.5.1) |
| bitops | bitops | 1.0-6 | 2013-08-17 | CRAN (R 3.5.1) |
| blob | blob | 1.2.1 | 2020-01-20 | CRAN (R 3.5.1) |
| brew | brew | 1.0-6 | 2011-04-13 | CRAN (R 3.5.1) |
| callr | callr | 3.4.2 | 2020-02-12 | CRAN (R 3.5.1) |
| caTools | caTools | 1.17.1.2 | 2019-03-06 | CRAN (R 3.5.1) |
| cli | cli | 2.0.2 | 2020-02-28 | CRAN (R 3.5.1) |
| cluster | cluster | 2.1.0 | 2019-06-19 | CRAN (R 3.5.1) |
| codetools | codetools | 0.2-16 | 2018-12-24 | CRAN (R 3.5.1) |
| colorspace | colorspace | 1.4-1 | 2019-03-18 | CRAN (R 3.5.1) |
| conos | conos | 1.3.0 | 2020-05-12 | local |
| cowplot | cowplot | 1.0.0 | 2019-07-11 | CRAN (R 3.5.1) |
| crayon | crayon | 1.3.4 | 2017-09-16 | CRAN (R 3.5.1) |
| data.table | data.table | 1.12.8 | 2019-12-09 | CRAN (R 3.5.1) |
| dataorganizer | dataorganizer | 0.1.0 | 2019-11-08 | local |
| DBI | DBI | 1.1.0 | 2019-12-15 | CRAN (R 3.5.1) |
| dendextend | dendextend | 1.13.4 | 2020-02-28 | CRAN (R 3.5.1) |
| desc | desc | 1.2.0 | 2018-05-01 | CRAN (R 3.5.1) |
| devtools | devtools | 2.2.2 | 2020-02-17 | CRAN (R 3.5.1) |
| digest | digest | 0.6.25 | 2020-02-23 | CRAN (R 3.5.1) |
| dplyr | dplyr | 0.8.5 | 2020-03-07 | CRAN (R 3.5.1) |
| dropestr | dropestr | 0.7.9 | 2019-02-02 | local |
| ellipsis | ellipsis | 0.3.0 | 2019-09-20 | CRAN (R 3.5.1) |
| Epilepsy19 | Epilepsy19 | 0.0.0.9000 | 2019-10-15 | local |
| evaluate | evaluate | 0.14 | 2019-05-28 | CRAN (R 3.5.1) |
| fansi | fansi | 0.4.1 | 2020-01-08 | CRAN (R 3.5.1) |
| farver | farver | 2.0.3 | 2020-01-16 | CRAN (R 3.5.1) |
| fastmap | fastmap | 1.0.1 | 2019-10-08 | CRAN (R 3.5.1) |
| fitdistrplus | fitdistrplus | 1.0-14 | 2019-01-23 | CRAN (R 3.5.1) |
| fs | fs | 1.3.2 | 2020-03-05 | CRAN (R 3.5.1) |
| future | future | 1.16.0 | 2020-01-16 | CRAN (R 3.5.1) |
| future.apply | future.apply | 1.4.0 | 2020-01-07 | CRAN (R 3.5.1) |
| gbRd | gbRd | 0.4-11 | 2012-10-01 | CRAN (R 3.5.1) |
| gdata | gdata | 2.18.0 | 2017-06-06 | CRAN (R 3.5.1) |
| ggbeeswarm | ggbeeswarm | 0.6.0 | 2018-10-16 | Github (eclarke/ggbeeswarm@fb85521) |
| ggplot2 | ggplot2 | 3.3.0 | 2020-03-05 | CRAN (R 3.5.1) |
| ggrastr | ggrastr | 0.1.7 | 2018-12-04 | Github (VPetukhov/ggrastr@203d5cc) |
| ggrepel | ggrepel | 0.8.2 | 2020-03-08 | CRAN (R 3.5.1) |
| ggridges | ggridges | 0.5.2 | 2020-01-12 | CRAN (R 3.5.1) |
| git2r | git2r | 0.26.1 | 2019-06-29 | CRAN (R 3.5.1) |
| globals | globals | 0.12.5 | 2019-12-07 | CRAN (R 3.5.1) |
| glue | glue | 1.3.2 | 2020-03-12 | CRAN (R 3.5.1) |
| gplots | gplots | 3.0.3 | 2020-02-25 | CRAN (R 3.5.1) |
| gridExtra | gridExtra | 2.3 | 2017-09-09 | CRAN (R 3.5.1) |
| gtable | gtable | 0.3.0 | 2019-03-25 | CRAN (R 3.5.1) |
| gtools | gtools | 3.8.1 | 2018-06-26 | CRAN (R 3.5.1) |
| hdf5r | hdf5r | 1.3.1 | 2020-01-10 | CRAN (R 3.5.1) |
| highr | highr | 0.8 | 2019-03-20 | CRAN (R 3.5.1) |
| htmltools | htmltools | 0.4.0 | 2019-10-04 | CRAN (R 3.5.1) |
| htmlwidgets | htmlwidgets | 1.5.1 | 2019-10-08 | CRAN (R 3.5.1) |
| httpuv | httpuv | 1.5.2 | 2019-09-11 | CRAN (R 3.5.1) |
| httr | httr | 1.4.1 | 2019-08-05 | CRAN (R 3.5.1) |
| ica | ica | 1.0-2 | 2018-05-24 | CRAN (R 3.5.1) |
| igraph | igraph | 1.2.4.2 | 2019-11-27 | CRAN (R 3.5.1) |
| IRanges | IRanges | 2.16.0 | 2018-10-30 | Bioconductor |
| irlba | irlba | 2.3.3 | 2019-02-05 | CRAN (R 3.5.1) |
| jsonlite | jsonlite | 1.6.1 | 2020-02-02 | CRAN (R 3.5.1) |
| KernSmooth | KernSmooth | 2.23-16 | 2019-10-15 | CRAN (R 3.5.1) |
| knitr | knitr | 1.28 | 2020-02-06 | CRAN (R 3.5.1) |
| labeling | labeling | 0.3 | 2014-08-23 | CRAN (R 3.5.1) |
| later | later | 1.0.0 | 2019-10-04 | CRAN (R 3.5.1) |
| lattice | lattice | 0.20-40 | 2020-02-19 | CRAN (R 3.5.1) |
| lazyeval | lazyeval | 0.2.2 | 2019-03-15 | CRAN (R 3.5.1) |
| leiden | leiden | 0.3.3 | 2020-02-04 | CRAN (R 3.5.1) |
| lifecycle | lifecycle | 0.2.0 | 2020-03-06 | CRAN (R 3.5.1) |
| listenv | listenv | 0.8.0 | 2019-12-05 | CRAN (R 3.5.1) |
| lmtest | lmtest | 0.9-37 | 2019-04-30 | CRAN (R 3.5.1) |
| lsei | lsei | 1.2-0 | 2017-10-23 | CRAN (R 3.5.1) |
| magrittr | magrittr | 1.5 | 2014-11-22 | CRAN (R 3.5.1) |
| MASS | MASS | 7.3-51.5 | 2019-12-20 | CRAN (R 3.5.1) |
| Matrix | Matrix | 1.2-18 | 2019-11-27 | CRAN (R 3.5.1) |
| memoise | memoise | 1.1.0 | 2017-04-21 | CRAN (R 3.5.1) |
| metap | metap | 1.3 | 2020-01-23 | CRAN (R 3.5.1) |
| mime | mime | 0.9 | 2020-02-04 | CRAN (R 3.5.1) |
| mnormt | mnormt | 1.5-6 | 2020-02-03 | CRAN (R 3.5.1) |
| multcomp | multcomp | 1.4-12 | 2020-01-10 | CRAN (R 3.5.1) |
| multtest | multtest | 2.38.0 | 2018-10-30 | Bioconductor |
| munsell | munsell | 0.5.0 | 2018-06-12 | CRAN (R 3.5.1) |
| mutoss | mutoss | 0.1-12 | 2017-12-04 | CRAN (R 3.5.1) |
| mvtnorm | mvtnorm | 1.1-0 | 2020-02-24 | CRAN (R 3.5.1) |
| nlme | nlme | 3.1-145 | 2020-03-04 | CRAN (R 3.5.1) |
| npsurv | npsurv | 0.4-0 | 2017-10-14 | CRAN (R 3.5.1) |
| numDeriv | numDeriv | 2016.8-1.1 | 2019-06-06 | CRAN (R 3.5.1) |
| org.Hs.eg.db | org.Hs.eg.db | 3.7.0 | 2019-10-08 | Bioconductor |
| pagoda2 | pagoda2 | 0.1.1 | 2019-12-10 | local |
| patchwork | patchwork | 1.0.0 | 2019-12-01 | CRAN (R 3.5.1) |
| pbapply | pbapply | 1.4-2 | 2019-08-31 | CRAN (R 3.5.1) |
| pillar | pillar | 1.4.3 | 2019-12-20 | CRAN (R 3.5.1) |
| pkgbuild | pkgbuild | 1.0.6 | 2019-10-09 | CRAN (R 3.5.1) |
| pkgconfig | pkgconfig | 2.0.3 | 2019-09-22 | CRAN (R 3.5.1) |
| pkgload | pkgload | 1.0.2 | 2018-10-29 | CRAN (R 3.5.1) |
| plotly | plotly | 4.9.2 | 2020-02-12 | CRAN (R 3.5.1) |
| plotrix | plotrix | 3.7-7 | 2019-12-05 | CRAN (R 3.5.1) |
| plyr | plyr | 1.8.6 | 2020-03-03 | CRAN (R 3.5.1) |
| png | png | 0.1-7 | 2013-12-03 | CRAN (R 3.5.1) |
| prettyunits | prettyunits | 1.1.1 | 2020-01-24 | CRAN (R 3.5.1) |
| processx | processx | 3.4.2 | 2020-02-09 | CRAN (R 3.5.1) |
| promises | promises | 1.1.0 | 2019-10-04 | CRAN (R 3.5.1) |
| ps | ps | 1.3.2 | 2020-02-13 | CRAN (R 3.5.1) |
| purrr | purrr | 0.3.3 | 2019-10-18 | CRAN (R 3.5.1) |
| R6 | R6 | 2.4.1 | 2019-11-12 | CRAN (R 3.5.1) |
| RANN | RANN | 2.6.1 | 2019-01-08 | CRAN (R 3.5.1) |
| rappdirs | rappdirs | 0.3.1 | 2016-03-28 | CRAN (R 3.5.1) |
| RColorBrewer | RColorBrewer | 1.1-2 | 2014-12-07 | CRAN (R 3.5.1) |
| Rcpp | Rcpp | 1.0.4 | 2020-03-17 | CRAN (R 3.5.1) |
| RcppAnnoy | RcppAnnoy | 0.0.16 | 2020-03-08 | CRAN (R 3.5.1) |
| Rdpack | Rdpack | 0.11-1 | 2019-12-14 | CRAN (R 3.5.1) |
| remotes | remotes | 2.1.1 | 2020-02-15 | CRAN (R 3.5.1) |
| reshape2 | reshape2 | 1.4.3 | 2017-12-11 | CRAN (R 3.5.1) |
| reticulate | reticulate | 1.14 | 2019-12-17 | CRAN (R 3.5.1) |
| rjson | rjson | 0.2.20 | 2018-06-08 | CRAN (R 3.5.1) |
| rlang | rlang | 0.4.5 | 2020-03-01 | CRAN (R 3.5.1) |
| rmarkdown | rmarkdown | 2.1 | 2020-01-20 | CRAN (R 3.5.1) |
| ROCR | ROCR | 1.0-7 | 2015-03-26 | CRAN (R 3.5.1) |
| Rook | Rook | 1.1-1 | 2014-10-20 | CRAN (R 3.5.1) |
| rprojroot | rprojroot | 1.3-2 | 2018-01-03 | CRAN (R 3.5.1) |
| RSQLite | RSQLite | 2.2.0 | 2020-01-07 | CRAN (R 3.5.1) |
| rstudioapi | rstudioapi | 0.11 | 2020-02-07 | CRAN (R 3.5.1) |
| rsvd | rsvd | 1.0.3 | 2020-02-17 | CRAN (R 3.5.1) |
| Rtsne | Rtsne | 0.15 | 2018-11-10 | CRAN (R 3.5.1) |
| S4Vectors | S4Vectors | 0.20.1 | 2018-11-09 | Bioconductor |
| sandwich | sandwich | 2.5-1 | 2019-04-06 | CRAN (R 3.5.1) |
| scales | scales | 1.1.0 | 2019-11-18 | CRAN (R 3.5.1) |
| sccore | sccore | 0.1 | 2020-04-24 | Github (hms-dbmi/sccore@2b34b61) |
| sctransform | sctransform | 0.2.1 | 2019-12-17 | CRAN (R 3.5.1) |
| sessioninfo | sessioninfo | 1.1.1 | 2018-11-05 | CRAN (R 3.5.1) |
| Seurat | Seurat | 3.1.4 | 2020-02-26 | CRAN (R 3.5.1) |
| shiny | shiny | 1.4.0.2 | 2020-03-13 | CRAN (R 3.5.1) |
| sn | sn | 1.5-5 | 2020-01-30 | CRAN (R 3.5.1) |
| stringi | stringi | 1.4.6 | 2020-02-17 | CRAN (R 3.5.1) |
| stringr | stringr | 1.4.0 | 2019-02-10 | CRAN (R 3.5.1) |
| survival | survival | 3.1-11 | 2020-03-07 | CRAN (R 3.5.1) |
| testthat | testthat | 2.3.2 | 2020-03-02 | CRAN (R 3.5.1) |
| TFisher | TFisher | 0.2.0 | 2018-03-21 | CRAN (R 3.5.1) |
| TH.data | TH.data | 1.0-10 | 2019-01-21 | CRAN (R 3.5.1) |
| tibble | tibble | 2.1.3 | 2019-06-06 | CRAN (R 3.5.1) |
| tidyr | tidyr | 1.0.2 | 2020-01-24 | CRAN (R 3.5.1) |
| tidyselect | tidyselect | 1.0.0 | 2020-01-27 | CRAN (R 3.5.1) |
| triebeard | triebeard | 0.3.0 | 2016-08-04 | CRAN (R 3.5.1) |
| tsne | tsne | 0.1-3 | 2016-07-15 | CRAN (R 3.5.1) |
| urltools | urltools | 1.7.3 | 2019-04-14 | CRAN (R 3.5.1) |
| usethis | usethis | 1.5.1 | 2019-07-04 | CRAN (R 3.5.1) |
| uwot | uwot | 0.1.8 | 2020-03-16 | CRAN (R 3.5.1) |
| vctrs | vctrs | 0.2.4 | 2020-03-10 | CRAN (R 3.5.1) |
| vipor | vipor | 0.4.5 | 2017-03-22 | CRAN (R 3.5.1) |
| viridis | viridis | 0.5.1 | 2018-03-29 | CRAN (R 3.5.1) |
| viridisLite | viridisLite | 0.3.0 | 2018-02-01 | CRAN (R 3.5.1) |
| whisker | whisker | 0.4 | 2019-08-28 | CRAN (R 3.5.1) |
| withr | withr | 2.1.2 | 2018-03-15 | CRAN (R 3.5.1) |
| workflowr | workflowr | 1.6.1 | 2020-03-11 | CRAN (R 3.5.1) |
| xfun | xfun | 0.12 | 2020-01-13 | CRAN (R 3.5.1) |
| xtable | xtable | 1.8-4 | 2019-04-21 | CRAN (R 3.5.1) |
| yaml | yaml | 2.2.1 | 2020-02-01 | CRAN (R 3.5.1) |
| zoo | zoo | 1.8-7 | 2020-01-10 | CRAN (R 3.5.1) |