Analysis of Smart-Seq2 data
08 July, 2020
Last updated: 2020-07-08
Checks: 7 0
Knit directory: Epilepsy19/
This reproducible R Markdown analysis was created with workflowr (version 1.6.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200706) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 856ddcf. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/fig_go.nb.html
Ignored: analysis/fig_neun.nb.html
Ignored: analysis/fig_overview.nb.html
Ignored: analysis/fig_smart_seq.nb.html
Ignored: analysis/fig_summary.nb.html
Ignored: analysis/fig_type_distance.nb.html
Ignored: analysis/gene_testing.nb.html
Ignored: analysis/prep_alignment.nb.html
Ignored: cache/con_allen.rds
Ignored: cache/con_filt_cells.rds
Ignored: cache/con_filt_samples.rds
Ignored: cache/con_ss.rds
Ignored: cache/count_matrices.rds
Ignored: cache/p2s/
Ignored: output/
Untracked files:
Untracked: DESCRIPTION
Untracked: NAMESPACE
Untracked: R/
Untracked: analysis/prep_alignment.Rmd
Untracked: analysis/prep_filtration.Rmd
Untracked: code/
Untracked: gene_modules/
Untracked: man/
Untracked: metadata/
Unstaged changes:
Modified: Epilepsy19.Rproj
Modified: README.md
Modified: analysis/index.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/fig_smart_seq.Rmd) and HTML (docs/fig_smart_seq.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5c8e634 | viktor_petukhov | 2020-07-08 | Fixed dates in fig_ notebooks |
| Rmd | 10f9bdc | viktor_petukhov | 2020-07-07 | Smart-seq notebook |
library(pagoda2)
library(conos)
library(magrittr)
library(ggplot2)
library(pbapply)
library(tidyverse)
library(cowplot)
library(readr)
library(scrattch.io)
library(ggalluvial)
devtools::load_all()
theme_set(theme_bw())
outPath <- function(...) OutputPath("fig_smart_seq", ...)
allenDataPath <- function(...) file.path("~/mh/Data/allen_human_cortex/", ...)Load data
con <- CachePath("con_filt_cells.rds") %>% read_rds()
sample_per_cell <- con$getDatasetPerCell()
annotation_by_level <- read_csv(MetadataPath("annotation.csv"))
annotation <- annotation_by_level %$% setNames(l4, cell) %>% .[names(sample_per_cell)] %>%
.[. != "Excluded"]
neuron_type_per_type <- ifelse(grepl("L[2-6].+", unique(annotation)), "Excitatory", "Inhibitory") %>%
setNames(unique(annotation))
type_order <- names(neuron_type_per_type)[order(neuron_type_per_type, names(neuron_type_per_type))]Alignment to our Smart-Seq dataset
so <- readRDS("/d0-mendel/home/demharters/R/projects/UPF9_14_17_19_22_23_24_32_33/seurat_upf.rds")
annot_old <- so@meta.data %$% setNames(subtypesKU, rownames(.))
cm_ss <- so@raw.data
dim(cm_ss)[1] 30632 955median(Matrix::colSums(cm_ss))[1] 1796751median(Matrix::colSums(cm_ss > 0))[1] 10302dim(cm_ss)[1] 30632 955Matrix::colSums(cm_ss > 0) %>% qplot(xlab="#Genes", ylab="#Cells")`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.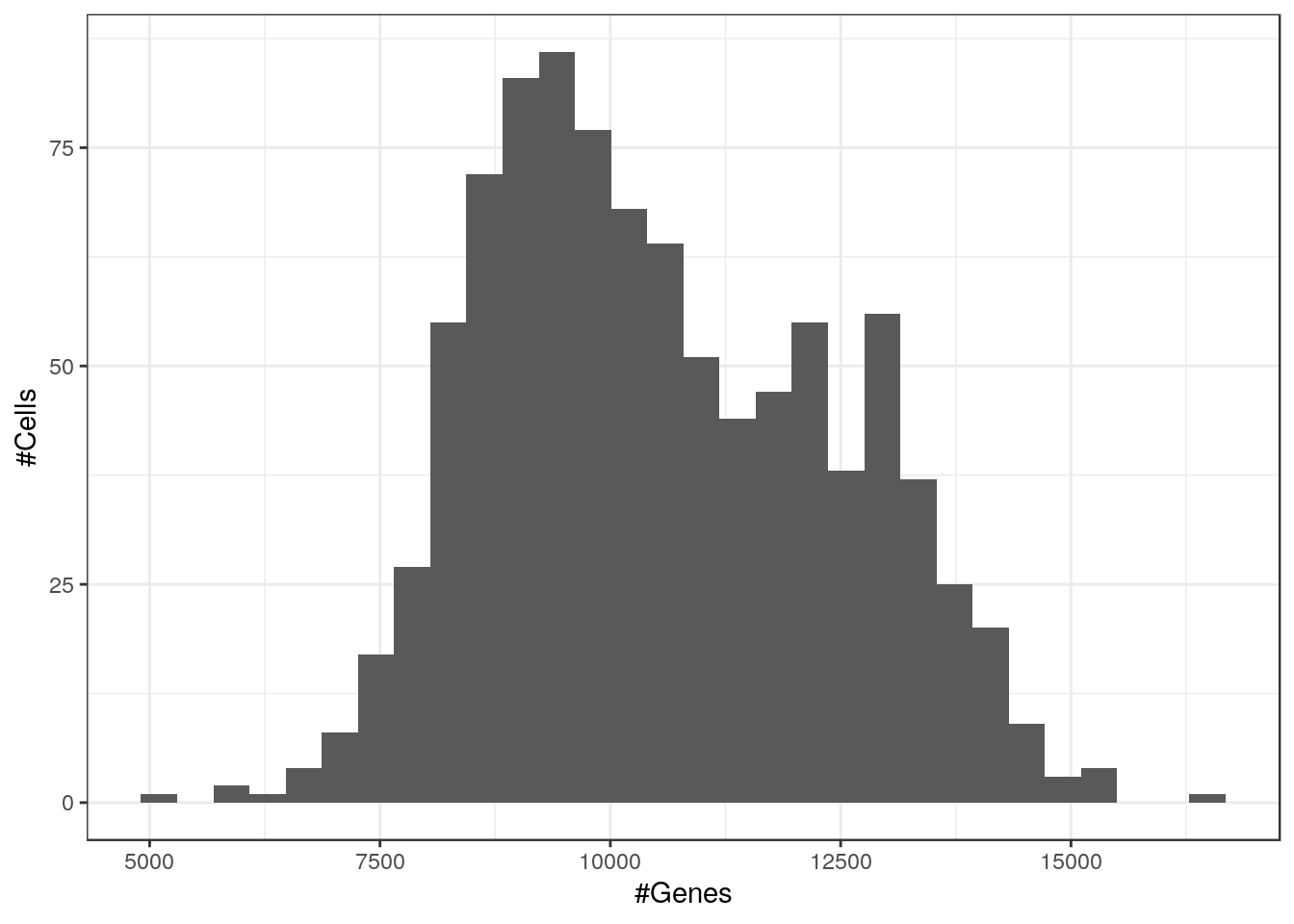
Matrix::colSums(cm_ss) %>% log10() %>% qplot()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.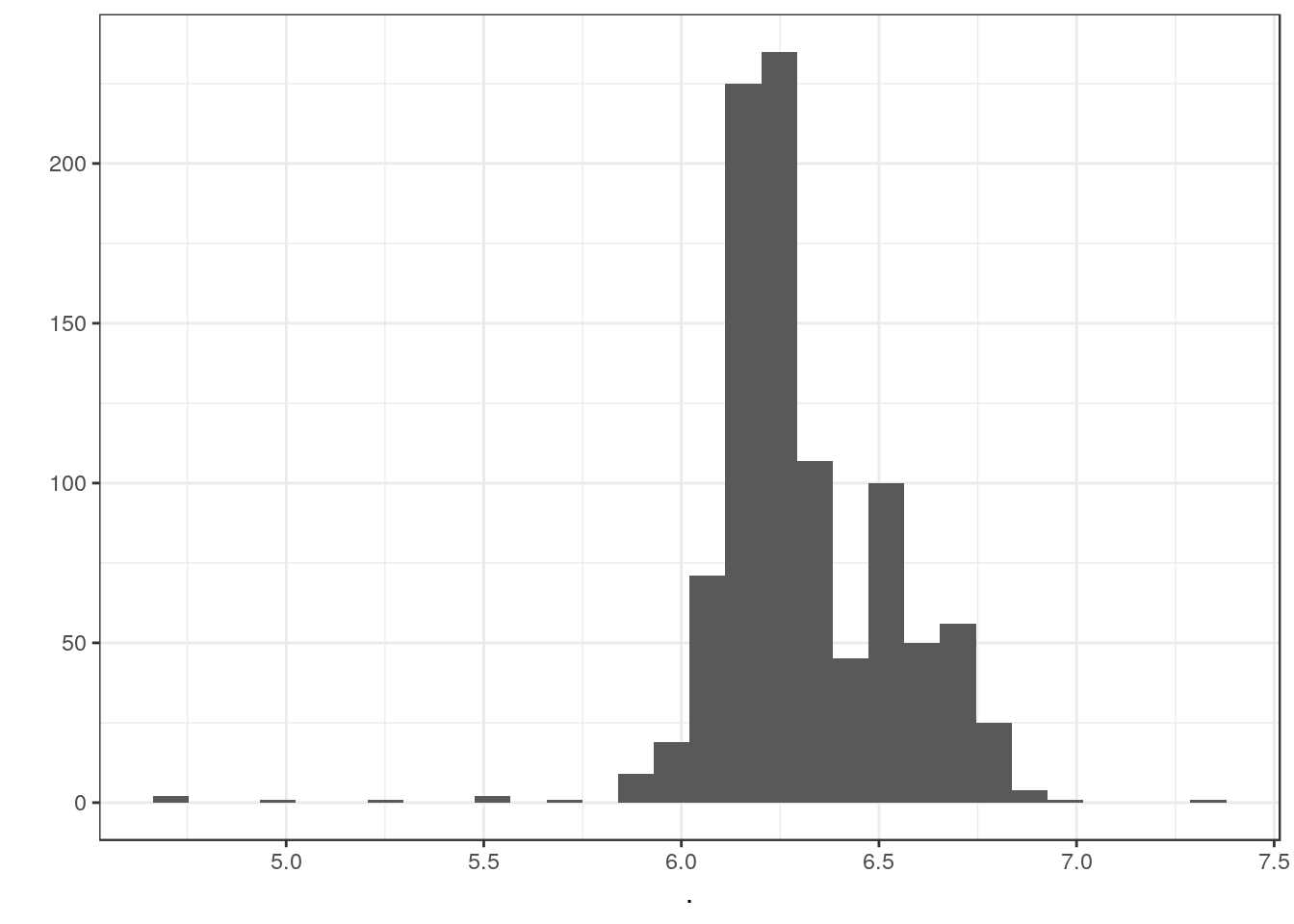
p2_ss <- GetPagoda(cm_ss)955 cells, 30632 genes; normalizing ... using plain model winsorizing ... log scale ... done.
calculating variance fit ... using gam 5012 overdispersed genes ... 5012persisting ... done.
running PCA using 1000 OD genes .... done
calculating distance ... pearson ...running tSNE using 30 cores:con_ss <- Conos$new(c(con$samples, list(SS2=p2_ss)), n.cores=40)
con_ss$buildGraph(verbose=T, var.scale=T, k=15, k.self=5, k.self.weight=0.1)found 0 out of 190 cached PCA space pairs ... running 190 additional PCA space pairs done
inter-sample links using mNN done
local pairs local pairs done
building graph ..donecon_ss$findCommunities(method=conos::leiden.community, resolution=10)
con_ss$embedGraph(method="UMAP", spread=1.5, min.dist=1, verbose=T, n.cores=30, min.prob.lower=1e-5)Convert graph to adjacency list...
Done
Estimate nearest neighbors and commute times...
Estimating hitting distances: 05:50:41.
Done.
Estimating commute distances: 05:50:52.
Hashing adjacency list: 05:50:52.
Done.
Estimating distances: 05:51:23.
Done
Done.
All done!: 05:51:47.
Done
Estimate UMAP embedding...05:51:48 UMAP embedding parameters a = 0.1034 b = 1.52405:51:48 Read 102937 rows and found 1 numeric columns05:51:50 Commencing smooth kNN distance calibration using 30 threads05:51:56 Initializing from normalized Laplacian + noise05:52:22 Commencing optimization for 1000 epochs, with 3928774 positive edges using 30 threads05:52:58 Optimization finishedDonewrite_rds(con_ss, CachePath("con_ss.rds"))
con_ss <- CachePath("con_ss.rds") %>% read_rds()annot_ss_new <- con_ss$propagateLabels(annotation, max.iters=50, verbose=T) %$%
labels[rownames(con_ss$samples$SS2$counts)]
tibble(Cell=names(annot_ss_new), Type=annot_ss_new) %>%
write_csv(MetadataPath("annotation_smart_seq.csv"))p_all <- con_ss$plotGraph(alpha=0.2, size=0.05, mark.groups=T, show.legend=F, groups=c(annotation, annot_ss_new),
raster=T, raster.dpi=150, font.size=4, shuffle.colors=T, show.labels=T, plot.na=F) +
theme(panel.grid=element_blank()) + labs(x="UMAP 1", y="UMAP 2")
p_emb <- con_ss$plotGraph(alpha=0.2, size=0.05, mark.groups=T, show.legend=F, groups=annot_ss_new,
raster=T, raster.dpi=150, font.size=4, show.labels=T) +
theme(panel.grid=element_blank()) + labs(x="UMAP 1", y="UMAP 2")
p_emb$layers[[3]]$aes_params$alpha <- 0.01
p_emb$layers[[1]]$aes_params$alpha <- 1
p_emb$layers[[2]] <- p_all$layers[[2]]
p_emb$layers <- p_emb$layers[c(3, 1, 2)]
p_emb$scales$scales[[2]] <- p_all$scales$scales[[2]]
# ggsave(outPath("ss_all.pdf"), p_all)
# ggsave(outPath("ss_subset.pdf"), p_emb)
p_all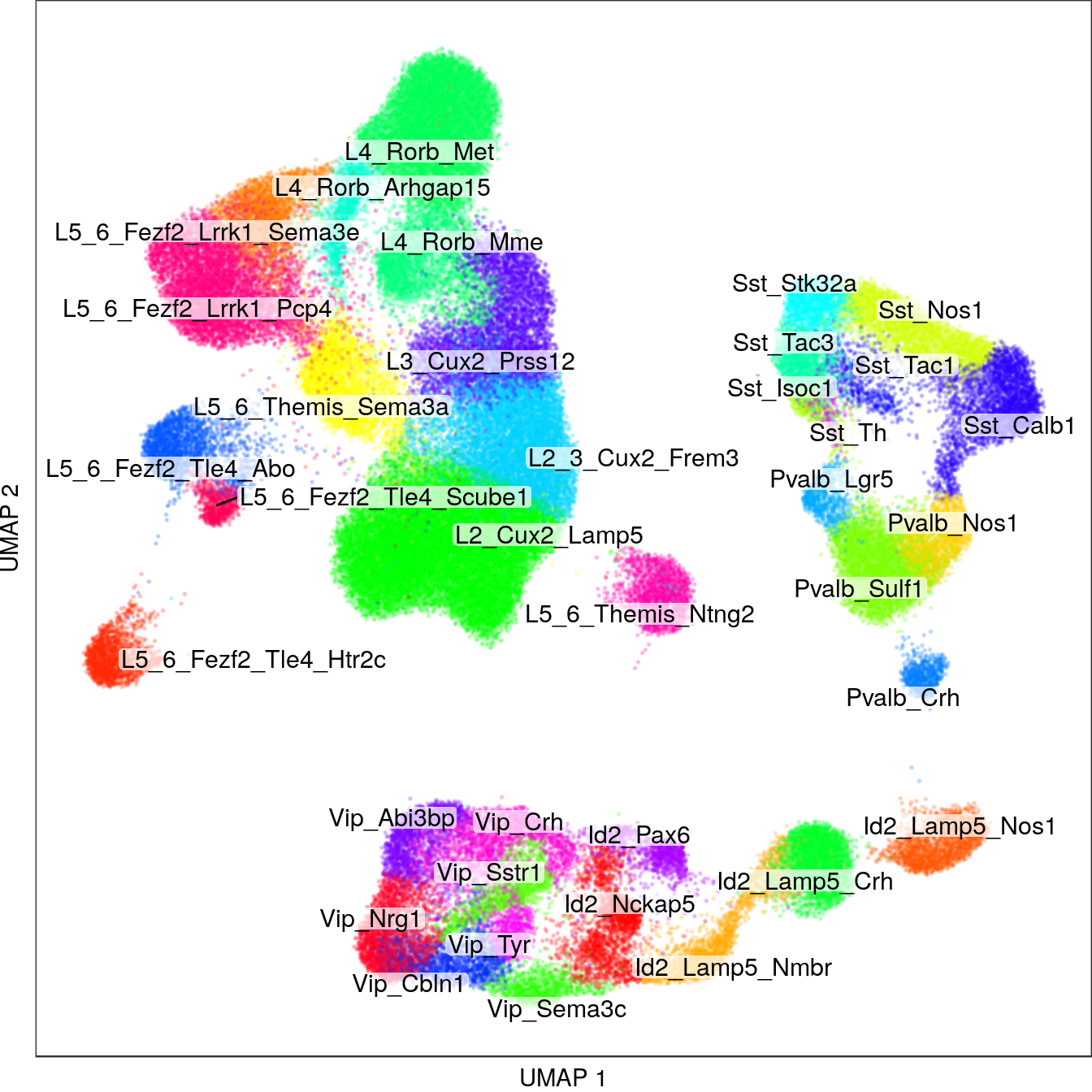
p_emb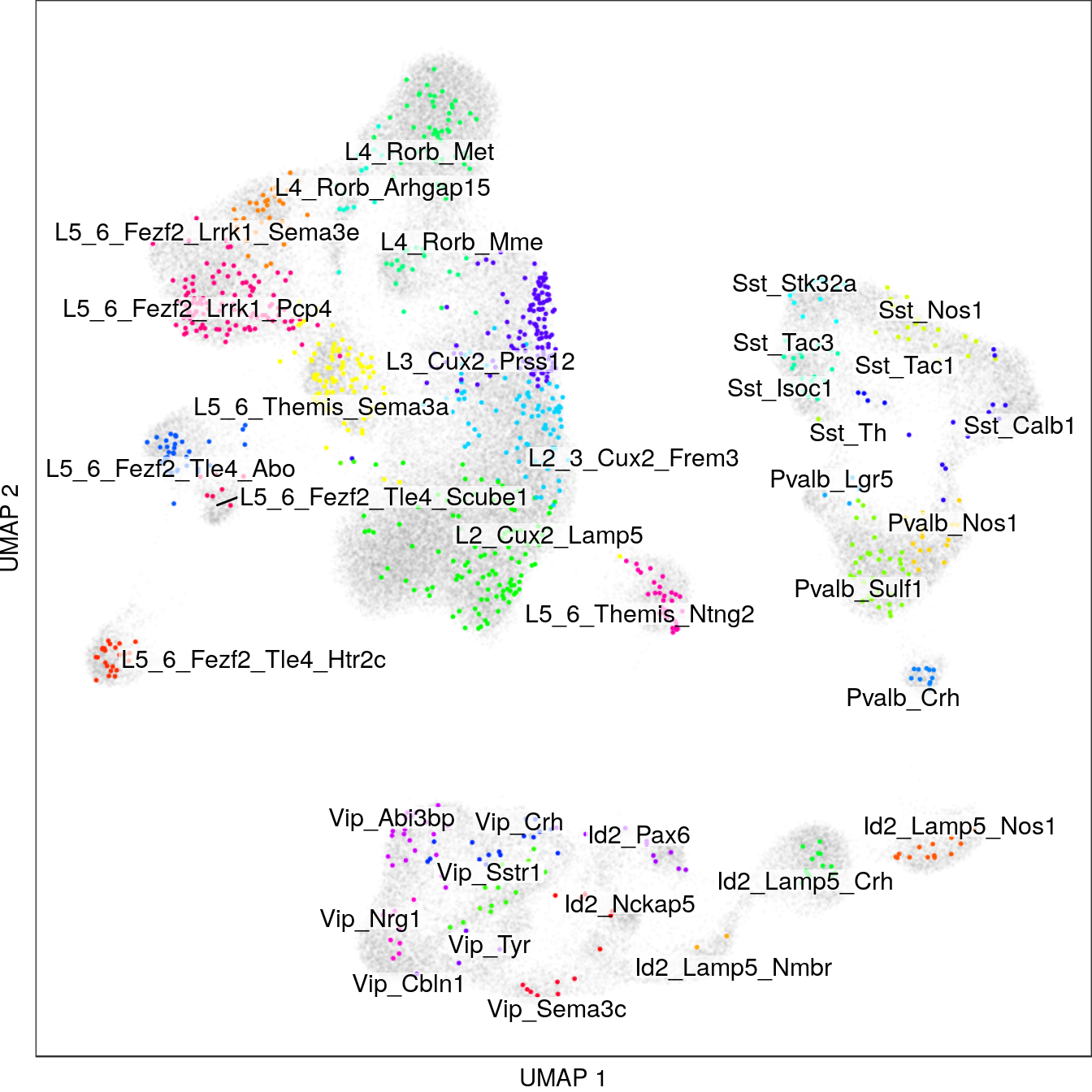
num_df <- table(Type=annot_ss_new) %>% as_tibble() %>%
mutate(Frac = n / sum(n), NeuronType=neuron_type_per_type[Type]) %>%
mutate(Type = factor(Type, levels=type_order))
ggplot(num_df) +
geom_bar(aes(x=Type, y=Frac * 100, fill=NeuronType), stat="identity") +
scale_y_continuous(expand=expansion(c(0, 0.05))) +
scale_fill_brewer("", palette="Set2") +
labs(x="", y="% of cells") +
theme(legend.position=c(1, 1.1), legend.justification=c(1, 1), panel.grid.major.x=element_blank(),
axis.text.x=element_text(angle=90, hjust=1, vjust=0.5), legend.background=element_blank())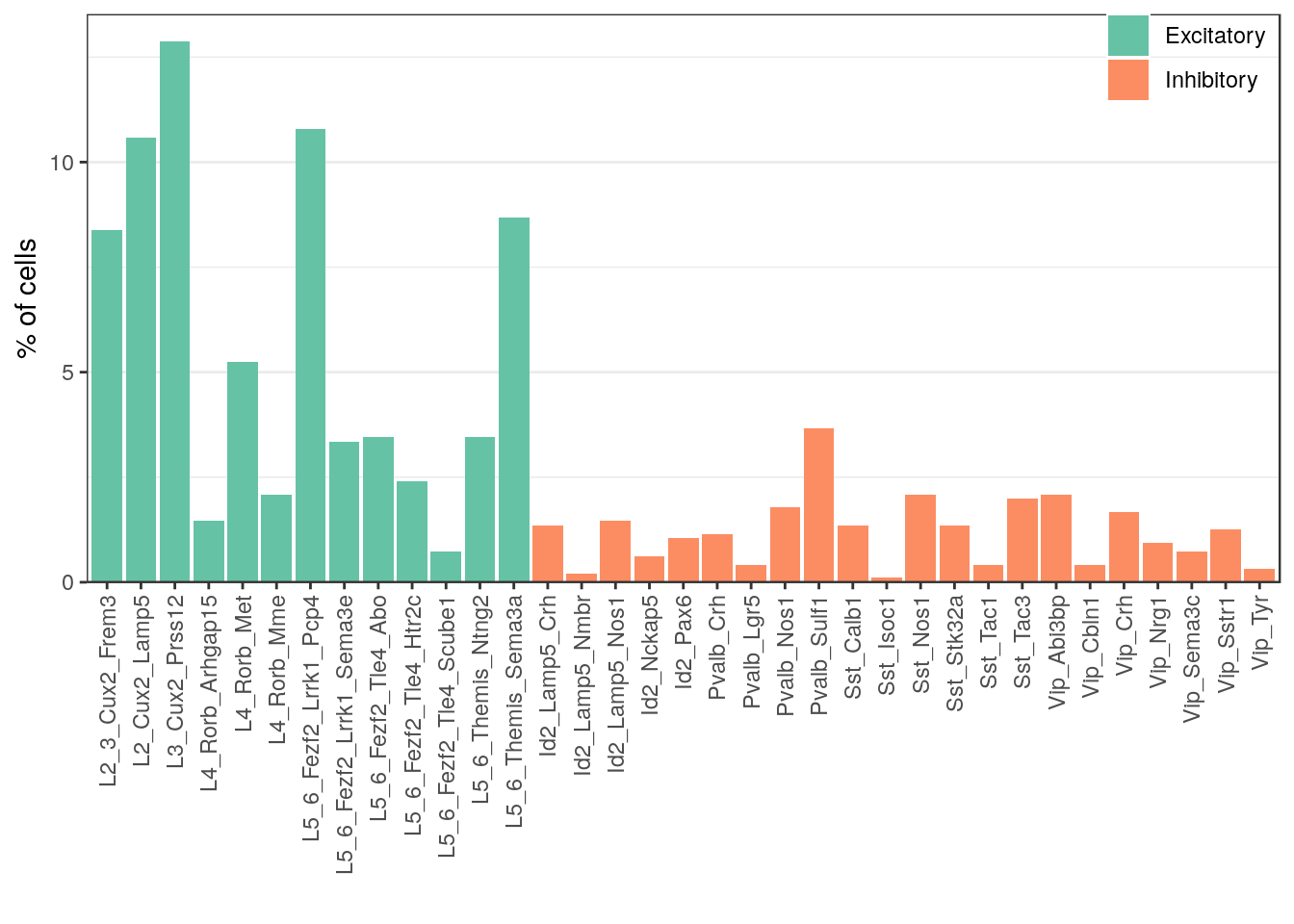
ggsave(outPath("ss_type_frac.pdf"))Annotation to Allen data
sample_info <- allenDataPath("sample_annotations.csv") %>% read_csv()
annot_allen <- sample_info %$% list(
l0=setNames(as.character(class_label), sample_name),
l1=setNames(as.character(subclass_label), sample_name),
l3=setNames(as.character(cluster_label), sample_name)
)
annot_allen$l2 <- strsplit(annot_allen$l3, " ") %>% sapply(function(x) paste(x[1:(length(x)-1)], collapse=" "))tome <- allenDataPath("transcrip.tome")
cm_allen <- read_tome_dgCMatrix(tome, "data/t_exon") %>%
set_colnames(read_tome_sample_names(tome)) %>% set_rownames(read_tome_gene_names(tome)) %>%
.[, !(annot_allen$l0[colnames(.)] %in% c("Exclude", "Non-neuronal"))]
dataset_id <- sample_info %$% setNames(external_donor_name_label, sample_name)
cms_per_dataset <- colnames(cm_allen) %>% split(dataset_id[.]) %>% lapply(function(ns) cm_allen[,ns])
p2s_allen <- lapply(cms_per_dataset, basicP2proc, n.cores=30, k=15,
get.largevis=F, make.geneknn=F, get.tsne=F)16248 cells, 50281 genes; normalizing ... using plain model winsorizing ... log scale ... done.
calculating variance fit ... using gam 13623 overdispersed genes ... 13623persisting ... done.
running PCA using 3000 OD genes .... done
9491 cells, 50281 genes; normalizing ... using plain model winsorizing ... log scale ... done.
calculating variance fit ... using gam 9971 overdispersed genes ... 9971persisting ... done.
running PCA using 3000 OD genes .... done
17013 cells, 50281 genes; normalizing ... using plain model winsorizing ... log scale ... done.
calculating variance fit ... using gam 14063 overdispersed genes ... 14063persisting ... done.
running PCA using 3000 OD genes .... donecon_allen <- con$samples[c("C6", "C7", "C8")] %>% c(p2s_allen) %>% Conos$new(n.cores=30)
source_fac <- names(con_allen$samples) %>% setNames(., .) %>% substr(1, 1)
con_allen$buildGraph(verbose=T, var.scale=T, k=20, k.self=5, k.self.weight=0.1, space="CCA",
same.factor.downweight=0.1, balancing.factor.per.sample=source_fac)found 0 out of 15 cached CCA space pairs ... running 15 additional CCA space pairs done
inter-sample links using mNN done
local pairs local pairs done
building graph ..done
balancing edge weights donecon_allen$embedGraph(method="UMAP", spread=1, min.dist=0.5, verbose=T, n.cores=30, min.prob.lower=1e-7)Convert graph to adjacency list...
Done
Estimate nearest neighbors and commute times...
Estimating hitting distances: 06:11:43.
Done.
Estimating commute distances: 06:11:50.
Hashing adjacency list: 06:11:50.
Done.
Estimating distances: 06:11:51.
Done
Done.
All done!: 06:12:02.
Done
Estimate UMAP embedding...
Donewrite_rds(con_allen, CachePath("con_allen.rds"))
con_allen <- CachePath("con_allen.rds") %>% read_rds()con_allen$plotGraph(color.by='sample', size=0.1, alpha=0.1, mark.groups=F, show.legend=T,
legend.pos=c(1, 1), raster=T, raster.dpi=150) +
theme(legend.title=element_blank())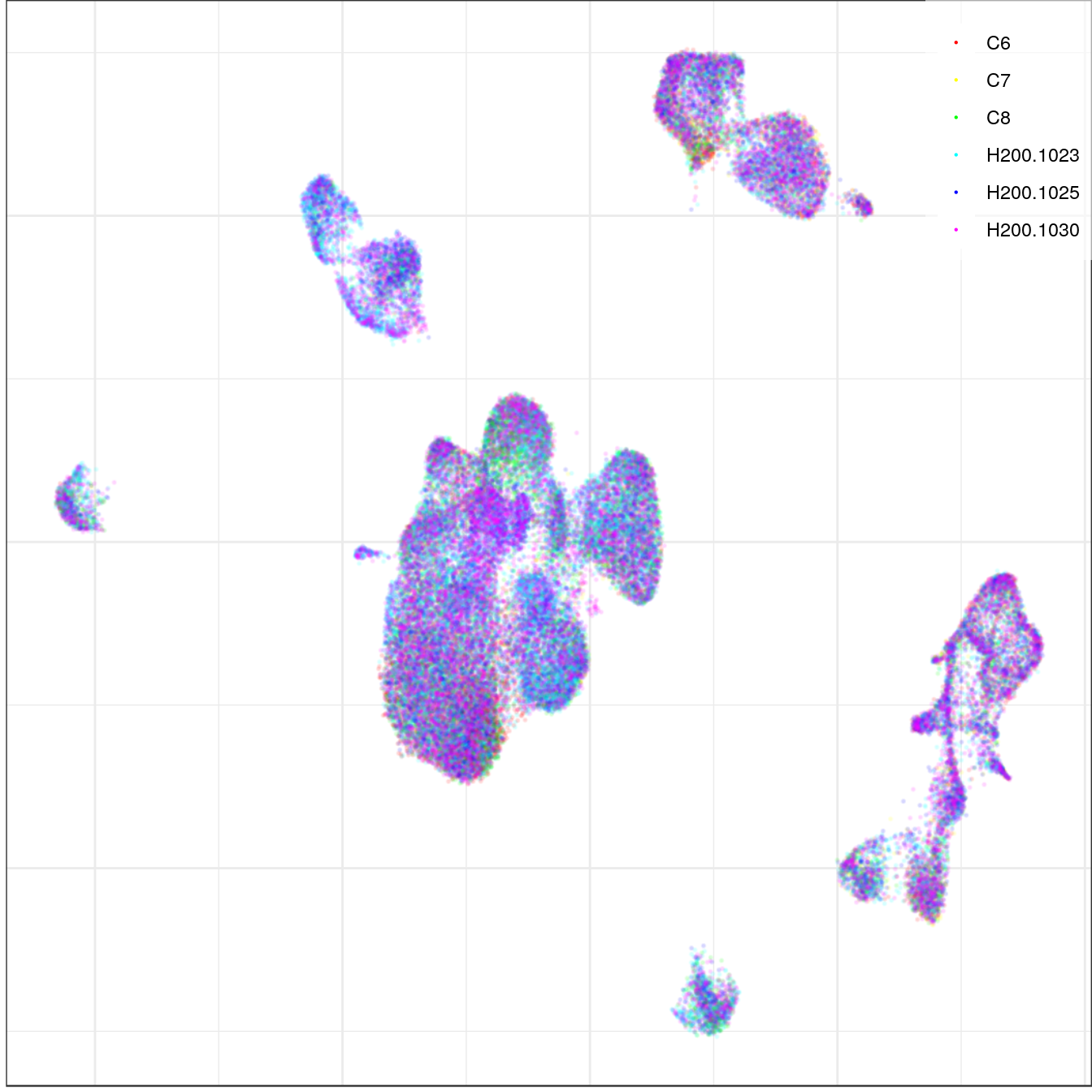
ggsave(outPath("allen_alignment_sample.pdf"))
con_allen$plotGraph(groups=annot_allen$l2, size=0.1, font.size=c(2, 3), alpha=0.1,
raster=T, raster.dpi=150)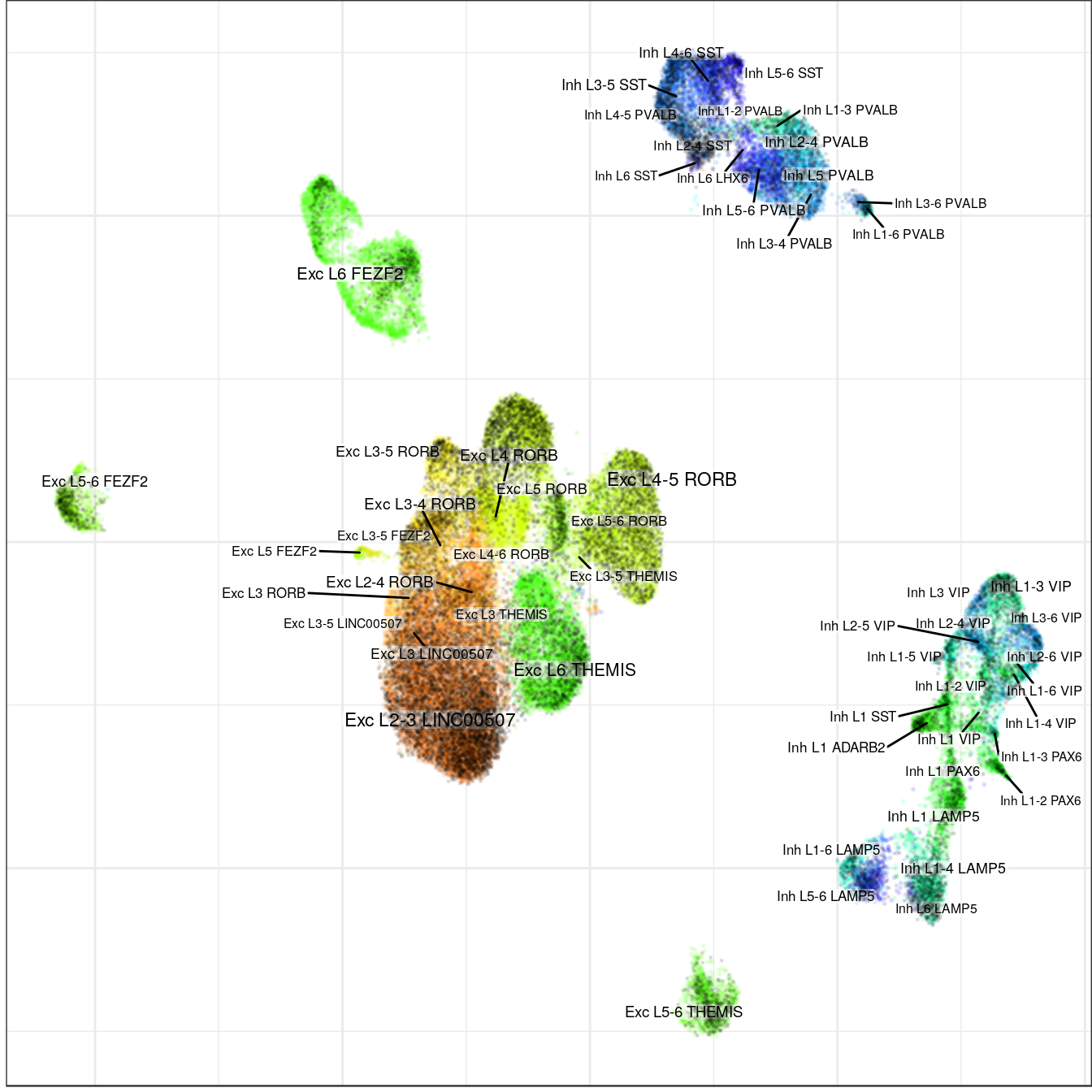
ggsave(outPath("allen_alignment_allen.pdf"))
con_allen$plotGraph(groups=annotation, size=0.1, font.size=c(2, 3), alpha=0.1,
raster=T, raster.dpi=150)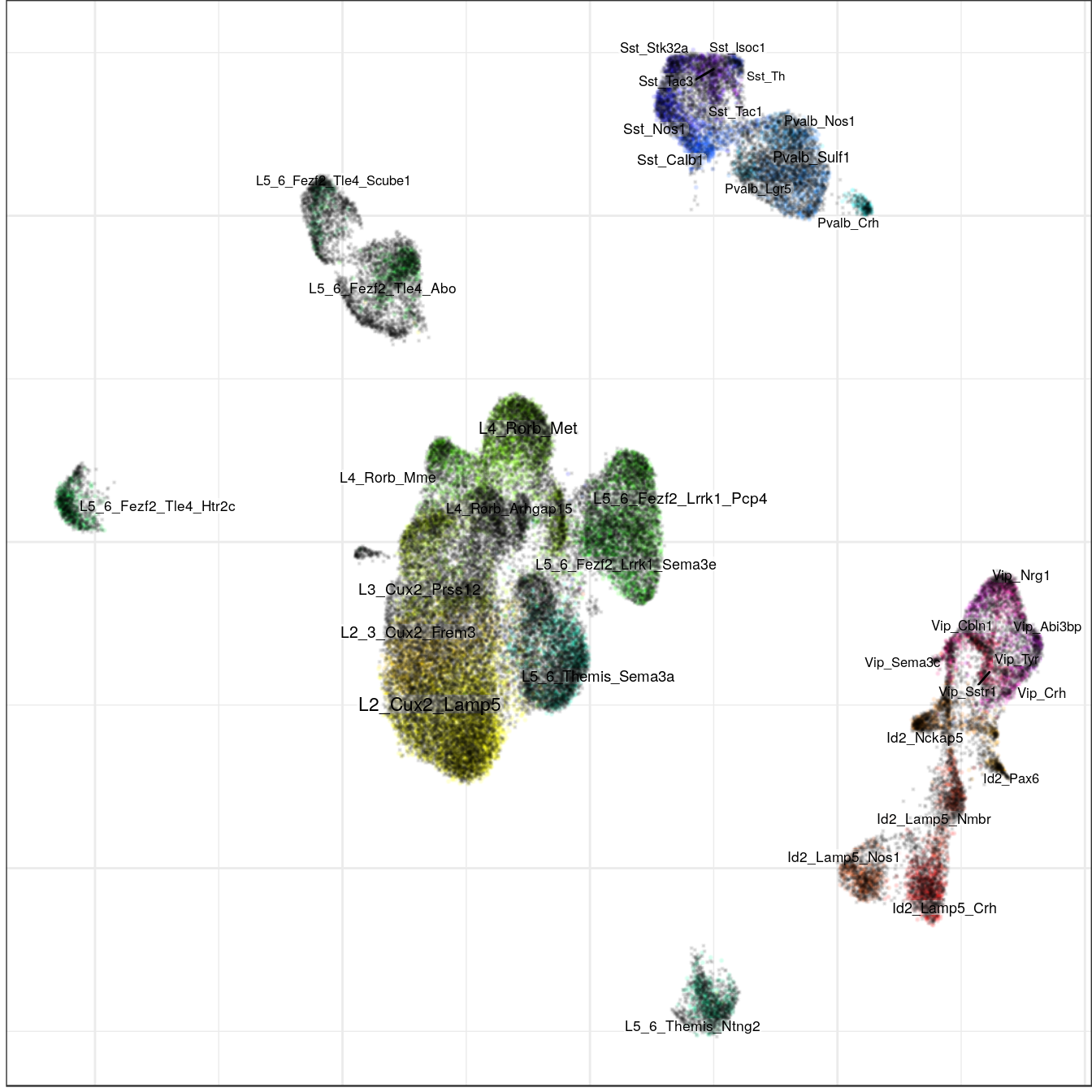
ggsave(outPath("allen_alignment_ours.pdf"))labels_prop <- con_allen$propagateLabels(annot_allen$l3, max.iters=50, verbose=T)
con_allen$plotGraph(groups=labels_prop$labels, size=0.1, font.size=c(2, 3), alpha=0.1)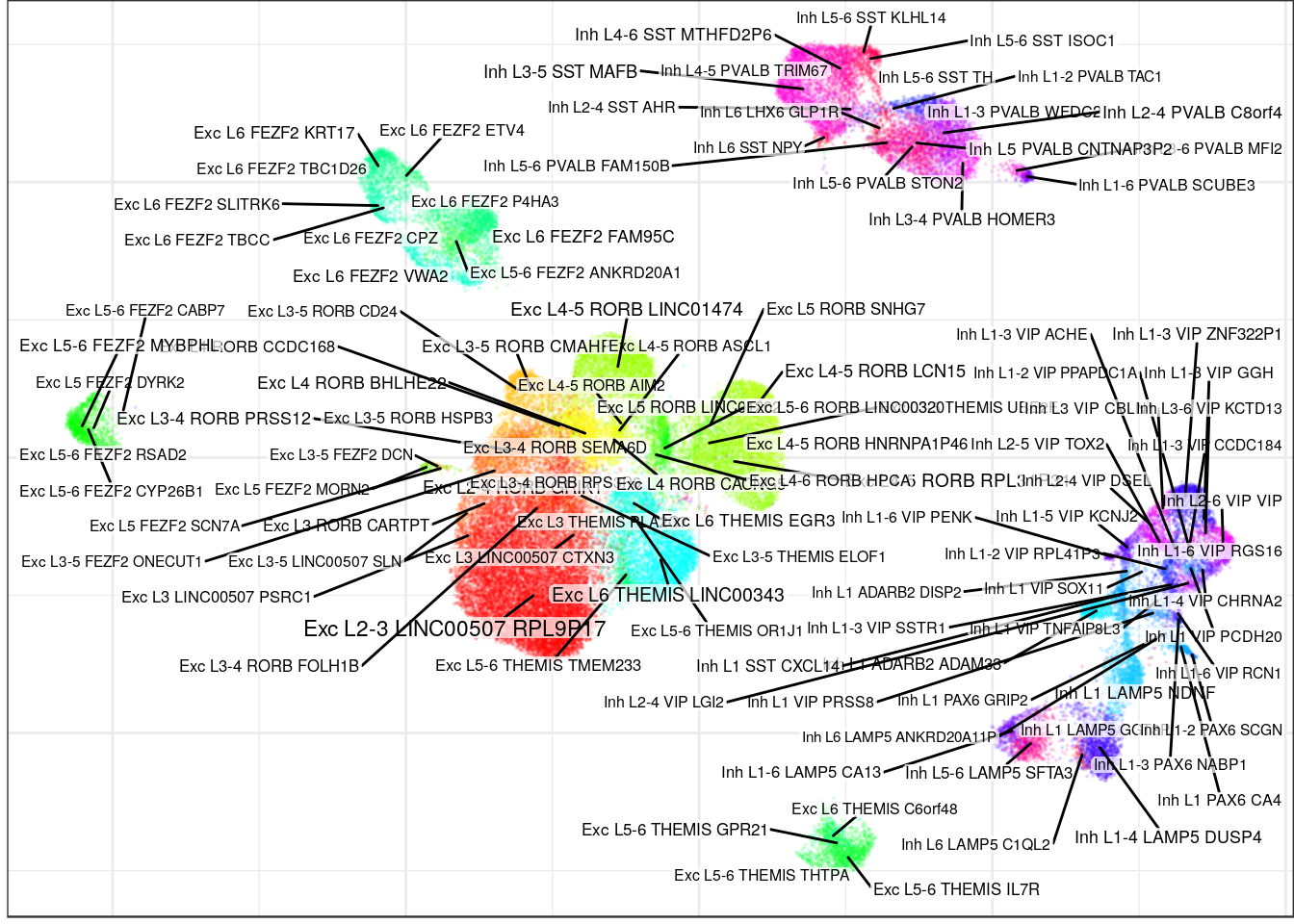
type_ranks <- factor(type_order, levels=type_order) %>% as.integer() %>% setNames(type_order)
t_ann <- annotation %>% .[intersect(names(.), names(con_allen$getDatasetPerCell()))]
freq_df <- table(KU=t_ann, Allen=labels_prop$labels[names(t_ann)]) %>% as_tibble() %>%
group_by(KU) %>% mutate(total_n=sum(n), freq=n / total_n) %>% ungroup() %>% filter(n > 1, freq > 0.05) %>%
mutate(NeuronType=neuron_type_per_type[KU])
allen_type_order <- freq_df %>% split(.$Allen) %>% lapply(function(x) x %$% setNames(n, KU) %>% `/`(sum(.))) %>%
sapply(function(ws) as.numeric(type_ranks[names(ws)[which.max(ws)]]) + sum(type_ranks[names(ws)] * ws) / 10) %>% sort() %>% names()
freq_df %<>% mutate(KU=factor(KU, levels=type_order), Allen=factor(Allen, levels=allen_type_order))plotAlluvium <- function(p.df, min.box.h, width.left, width.right, alpha=1.0, ...) {
s.ku <- p.df %$% split(n, droplevels(KU)) %>% sapply(sum)
s.allen <- p.df %$% split(n, droplevels(Allen)) %>% sapply(sum)
s.total <- c(rev(s.ku), rev(s.allen)) %>% sqrt() %>% sqrt() %>% `/`(max(.))
palette <- sccore::fac2col(p.df$KU, return.details=T, ...) %$%
setNames(sample(palette), names(palette)) %>% alpha(alpha=alpha)
ggplot(p.df, aes(axis1=KU, axis2=Allen, y=pmax(sqrt(n), min.box.h))) +
geom_alluvium(aes(fill=KU)) +
geom_stratum(width=c(rep(width.left, length(s.ku)), rep(width.right, length(s.allen)))) +
geom_text(stat="stratum", infer.label=TRUE, size=s.total * 2 + 1) +
scale_x_continuous(expand=c(0, 0)) +
scale_y_continuous(expand=c(0, 0)) +
theme_void() + theme(legend.position="none") +
scale_fill_manual(values=palette)
}filter(freq_df, NeuronType == "Excitatory") %>% plotAlluvium(10, 0.3, 0.5, v=0.7, s=1.0)
ggsave(outPath("allen_mapping_ex.pdf"))filter(freq_df, NeuronType == "Inhibitory") %>%
plotAlluvium(2, 0.25, 0.4, v=0.8, s=1.0)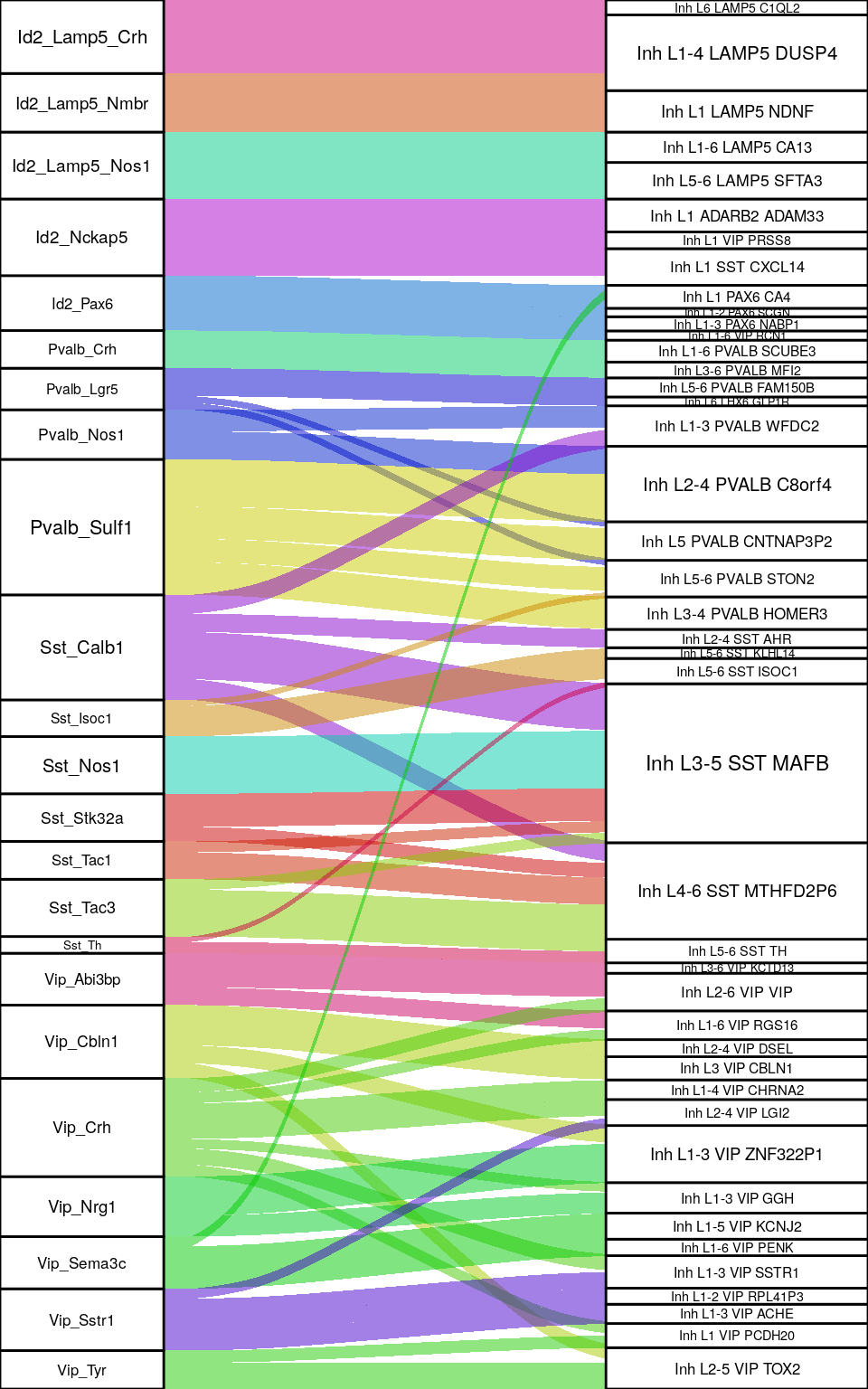
ggsave(outPath("allen_mapping_inh.pdf"))
data.frame(value=unlist(sessioninfo::platform_info()))| value | |
|---|---|
| version | R version 3.5.1 (2018-07-02) |
| os | Ubuntu 18.04.2 LTS |
| system | x86_64, linux-gnu |
| ui | X11 |
| language | (EN) |
| collate | en_US.UTF-8 |
| ctype | en_US.UTF-8 |
| tz | America/New_York |
| date | 2020-07-08 |
as.data.frame(sessioninfo::package_info())[c('package', 'loadedversion', 'date', 'source')]| package | loadedversion | date | source | |
|---|---|---|---|---|
| AnnotationDbi | AnnotationDbi | 1.44.0 | 2018-10-30 | Bioconductor |
| assertthat | assertthat | 0.2.1 | 2019-03-21 | CRAN (R 3.5.1) |
| backports | backports | 1.1.5 | 2019-10-02 | CRAN (R 3.5.1) |
| base64enc | base64enc | 0.1-3 | 2015-07-28 | CRAN (R 3.5.1) |
| beeswarm | beeswarm | 0.2.3 | 2016-04-25 | CRAN (R 3.5.1) |
| Biobase | Biobase | 2.42.0 | 2018-10-30 | Bioconductor |
| BiocGenerics | BiocGenerics | 0.28.0 | 2018-10-30 | Bioconductor |
| bit | bit | 1.1-15.2 | 2020-02-10 | CRAN (R 3.5.1) |
| bit64 | bit64 | 0.9-7 | 2017-05-08 | CRAN (R 3.5.1) |
| blob | blob | 1.2.1 | 2020-01-20 | CRAN (R 3.5.1) |
| brew | brew | 1.0-6 | 2011-04-13 | CRAN (R 3.5.1) |
| broom | broom | 0.5.5 | 2020-02-29 | CRAN (R 3.5.1) |
| Cairo | Cairo | 1.5-11 | 2020-03-09 | CRAN (R 3.5.1) |
| callr | callr | 3.4.2 | 2020-02-12 | CRAN (R 3.5.1) |
| cellranger | cellranger | 1.1.0 | 2016-07-27 | CRAN (R 3.5.1) |
| cli | cli | 2.0.2 | 2020-02-28 | CRAN (R 3.5.1) |
| colorspace | colorspace | 1.4-1 | 2019-03-18 | CRAN (R 3.5.1) |
| conos | conos | 1.3.0 | 2020-05-12 | local |
| cowplot | cowplot | 1.0.0 | 2019-07-11 | CRAN (R 3.5.1) |
| crayon | crayon | 1.3.4 | 2017-09-16 | CRAN (R 3.5.1) |
| data.table | data.table | 1.12.8 | 2019-12-09 | CRAN (R 3.5.1) |
| dataorganizer | dataorganizer | 0.1.0 | 2019-11-08 | local |
| DBI | DBI | 1.1.0 | 2019-12-15 | CRAN (R 3.5.1) |
| dbplyr | dbplyr | 1.4.2 | 2019-06-17 | CRAN (R 3.5.1) |
| dendextend | dendextend | 1.13.4 | 2020-02-28 | CRAN (R 3.5.1) |
| desc | desc | 1.2.0 | 2018-05-01 | CRAN (R 3.5.1) |
| devtools | devtools | 2.2.2 | 2020-02-17 | CRAN (R 3.5.1) |
| digest | digest | 0.6.25 | 2020-02-23 | CRAN (R 3.5.1) |
| dplyr | dplyr | 0.8.5 | 2020-03-07 | CRAN (R 3.5.1) |
| ellipsis | ellipsis | 0.3.0 | 2019-09-20 | CRAN (R 3.5.1) |
| Epilepsy19 | Epilepsy19 | 0.0.0.9000 | 2019-10-15 | local |
| evaluate | evaluate | 0.14 | 2019-05-28 | CRAN (R 3.5.1) |
| fansi | fansi | 0.4.1 | 2020-01-08 | CRAN (R 3.5.1) |
| farver | farver | 2.0.3 | 2020-01-16 | CRAN (R 3.5.1) |
| fastmap | fastmap | 1.0.1 | 2019-10-08 | CRAN (R 3.5.1) |
| forcats | forcats | 0.5.0 | 2020-03-01 | CRAN (R 3.5.1) |
| fs | fs | 1.3.2 | 2020-03-05 | CRAN (R 3.5.1) |
| generics | generics | 0.0.2 | 2018-11-29 | CRAN (R 3.5.1) |
| ggalluvial | ggalluvial | 0.11.1 | 2019-12-03 | CRAN (R 3.5.1) |
| ggbeeswarm | ggbeeswarm | 0.6.0 | 2018-10-16 | Github (eclarke/ggbeeswarm@fb85521) |
| ggplot2 | ggplot2 | 3.3.0 | 2020-03-05 | CRAN (R 3.5.1) |
| ggrastr | ggrastr | 0.1.7 | 2018-12-04 | Github (VPetukhov/ggrastr@203d5cc) |
| ggrepel | ggrepel | 0.8.2 | 2020-03-08 | CRAN (R 3.5.1) |
| git2r | git2r | 0.26.1 | 2019-06-29 | CRAN (R 3.5.1) |
| glue | glue | 1.3.2 | 2020-03-12 | CRAN (R 3.5.1) |
| gridExtra | gridExtra | 2.3 | 2017-09-09 | CRAN (R 3.5.1) |
| gtable | gtable | 0.3.0 | 2019-03-25 | CRAN (R 3.5.1) |
| haven | haven | 2.2.0 | 2019-11-08 | CRAN (R 3.5.1) |
| highr | highr | 0.8 | 2019-03-20 | CRAN (R 3.5.1) |
| hms | hms | 0.5.3 | 2020-01-08 | CRAN (R 3.5.1) |
| htmltools | htmltools | 0.4.0 | 2019-10-04 | CRAN (R 3.5.1) |
| httpuv | httpuv | 1.5.2 | 2019-09-11 | CRAN (R 3.5.1) |
| httr | httr | 1.4.1 | 2019-08-05 | CRAN (R 3.5.1) |
| igraph | igraph | 1.2.4.2 | 2019-11-27 | CRAN (R 3.5.1) |
| IRanges | IRanges | 2.16.0 | 2018-10-30 | Bioconductor |
| irlba | irlba | 2.3.3 | 2019-02-05 | CRAN (R 3.5.1) |
| jsonlite | jsonlite | 1.6.1 | 2020-02-02 | CRAN (R 3.5.1) |
| knitr | knitr | 1.28 | 2020-02-06 | CRAN (R 3.5.1) |
| labeling | labeling | 0.3 | 2014-08-23 | CRAN (R 3.5.1) |
| later | later | 1.0.0 | 2019-10-04 | CRAN (R 3.5.1) |
| lattice | lattice | 0.20-40 | 2020-02-19 | CRAN (R 3.5.1) |
| lazyeval | lazyeval | 0.2.2 | 2019-03-15 | CRAN (R 3.5.1) |
| lifecycle | lifecycle | 0.2.0 | 2020-03-06 | CRAN (R 3.5.1) |
| lubridate | lubridate | 1.7.4 | 2018-04-11 | CRAN (R 3.5.1) |
| magrittr | magrittr | 1.5 | 2014-11-22 | CRAN (R 3.5.1) |
| MASS | MASS | 7.3-51.5 | 2019-12-20 | CRAN (R 3.5.1) |
| Matrix | Matrix | 1.2-18 | 2019-11-27 | CRAN (R 3.5.1) |
| memoise | memoise | 1.1.0 | 2017-04-21 | CRAN (R 3.5.1) |
| mgcv | mgcv | 1.8-31 | 2019-11-09 | CRAN (R 3.5.1) |
| mime | mime | 0.9 | 2020-02-04 | CRAN (R 3.5.1) |
| modelr | modelr | 0.1.6 | 2020-02-22 | CRAN (R 3.5.1) |
| munsell | munsell | 0.5.0 | 2018-06-12 | CRAN (R 3.5.1) |
| nlme | nlme | 3.1-145 | 2020-03-04 | CRAN (R 3.5.1) |
| org.Hs.eg.db | org.Hs.eg.db | 3.7.0 | 2019-10-08 | Bioconductor |
| pagoda2 | pagoda2 | 0.1.1 | 2019-12-10 | local |
| pbapply | pbapply | 1.4-2 | 2019-08-31 | CRAN (R 3.5.1) |
| pillar | pillar | 1.4.3 | 2019-12-20 | CRAN (R 3.5.1) |
| pkgbuild | pkgbuild | 1.0.6 | 2019-10-09 | CRAN (R 3.5.1) |
| pkgconfig | pkgconfig | 2.0.3 | 2019-09-22 | CRAN (R 3.5.1) |
| pkgload | pkgload | 1.0.2 | 2018-10-29 | CRAN (R 3.5.1) |
| plyr | plyr | 1.8.6 | 2020-03-03 | CRAN (R 3.5.1) |
| prettyunits | prettyunits | 1.1.1 | 2020-01-24 | CRAN (R 3.5.1) |
| processx | processx | 3.4.2 | 2020-02-09 | CRAN (R 3.5.1) |
| promises | promises | 1.1.0 | 2019-10-04 | CRAN (R 3.5.1) |
| ps | ps | 1.3.2 | 2020-02-13 | CRAN (R 3.5.1) |
| purrr | purrr | 0.3.3 | 2019-10-18 | CRAN (R 3.5.1) |
| R6 | R6 | 2.4.1 | 2019-11-12 | CRAN (R 3.5.1) |
| RColorBrewer | RColorBrewer | 1.1-2 | 2014-12-07 | CRAN (R 3.5.1) |
| Rcpp | Rcpp | 1.0.4 | 2020-03-17 | CRAN (R 3.5.1) |
| readr | readr | 1.3.1 | 2018-12-21 | CRAN (R 3.5.1) |
| readxl | readxl | 1.3.1 | 2019-03-13 | CRAN (R 3.5.1) |
| remotes | remotes | 2.1.1 | 2020-02-15 | CRAN (R 3.5.1) |
| reprex | reprex | 0.3.0 | 2019-05-16 | CRAN (R 3.5.1) |
| rhdf5 | rhdf5 | 2.26.2 | 2019-01-02 | Bioconductor |
| Rhdf5lib | Rhdf5lib | 1.4.3 | 2019-03-25 | Bioconductor |
| rjson | rjson | 0.2.20 | 2018-06-08 | CRAN (R 3.5.1) |
| rlang | rlang | 0.4.5 | 2020-03-01 | CRAN (R 3.5.1) |
| rmarkdown | rmarkdown | 2.1 | 2020-01-20 | CRAN (R 3.5.1) |
| Rook | Rook | 1.1-1 | 2014-10-20 | CRAN (R 3.5.1) |
| rprojroot | rprojroot | 1.3-2 | 2018-01-03 | CRAN (R 3.5.1) |
| RSpectra | RSpectra | 0.16-0 | 2019-12-01 | CRAN (R 3.5.1) |
| RSQLite | RSQLite | 2.2.0 | 2020-01-07 | CRAN (R 3.5.1) |
| rstudioapi | rstudioapi | 0.11 | 2020-02-07 | CRAN (R 3.5.1) |
| Rtsne | Rtsne | 0.15 | 2018-11-10 | CRAN (R 3.5.1) |
| rvest | rvest | 0.3.5 | 2019-11-08 | CRAN (R 3.5.1) |
| S4Vectors | S4Vectors | 0.20.1 | 2018-11-09 | Bioconductor |
| scales | scales | 1.1.0 | 2019-11-18 | CRAN (R 3.5.1) |
| sccore | sccore | 0.1 | 2020-04-24 | Github (hms-dbmi/sccore@2b34b61) |
| scrattch.io | scrattch.io | 0.1.0 | 2019-10-16 | Github (AllenInstitute/scrattch.io@11ca1e0) |
| sessioninfo | sessioninfo | 1.1.1 | 2018-11-05 | CRAN (R 3.5.1) |
| shiny | shiny | 1.4.0.2 | 2020-03-13 | CRAN (R 3.5.1) |
| stringi | stringi | 1.4.6 | 2020-02-17 | CRAN (R 3.5.1) |
| stringr | stringr | 1.4.0 | 2019-02-10 | CRAN (R 3.5.1) |
| testthat | testthat | 2.3.2 | 2020-03-02 | CRAN (R 3.5.1) |
| tibble | tibble | 2.1.3 | 2019-06-06 | CRAN (R 3.5.1) |
| tidyr | tidyr | 1.0.2 | 2020-01-24 | CRAN (R 3.5.1) |
| tidyselect | tidyselect | 1.0.0 | 2020-01-27 | CRAN (R 3.5.1) |
| tidyverse | tidyverse | 1.3.0 | 2019-11-21 | CRAN (R 3.5.1) |
| triebeard | triebeard | 0.3.0 | 2016-08-04 | CRAN (R 3.5.1) |
| urltools | urltools | 1.7.3 | 2019-04-14 | CRAN (R 3.5.1) |
| usethis | usethis | 1.5.1 | 2019-07-04 | CRAN (R 3.5.1) |
| uwot | uwot | 0.1.8 | 2020-03-16 | CRAN (R 3.5.1) |
| vctrs | vctrs | 0.2.4 | 2020-03-10 | CRAN (R 3.5.1) |
| vipor | vipor | 0.4.5 | 2017-03-22 | CRAN (R 3.5.1) |
| viridis | viridis | 0.5.1 | 2018-03-29 | CRAN (R 3.5.1) |
| viridisLite | viridisLite | 0.3.0 | 2018-02-01 | CRAN (R 3.5.1) |
| whisker | whisker | 0.4 | 2019-08-28 | CRAN (R 3.5.1) |
| withr | withr | 2.1.2 | 2018-03-15 | CRAN (R 3.5.1) |
| workflowr | workflowr | 1.6.1 | 2020-03-11 | CRAN (R 3.5.1) |
| xfun | xfun | 0.12 | 2020-01-13 | CRAN (R 3.5.1) |
| xml2 | xml2 | 1.2.5 | 2020-03-11 | CRAN (R 3.5.1) |
| xtable | xtable | 1.8-4 | 2019-04-21 | CRAN (R 3.5.1) |
| yaml | yaml | 2.2.1 | 2020-02-01 | CRAN (R 3.5.1) |